 There is no doubt that the little bugs that live inside our bodies and on our skin affect our health and well-being, even our state of mind, but what we want to know is, by studying them, can we prevent disease or even just a bad mood? And how can we monitor them on a daily basis? Will there be an app called “What’s up microbes?” that communicates with some new device that checks on those microbes and reports back what we should eat or if we should go see the doctor, or go for a run, or take it easy and relax to ensure we maintain a healthy microbial equilibrium?
There is no doubt that the little bugs that live inside our bodies and on our skin affect our health and well-being, even our state of mind, but what we want to know is, by studying them, can we prevent disease or even just a bad mood? And how can we monitor them on a daily basis? Will there be an app called “What’s up microbes?” that communicates with some new device that checks on those microbes and reports back what we should eat or if we should go see the doctor, or go for a run, or take it easy and relax to ensure we maintain a healthy microbial equilibrium?
A few months ago, I shipped my “gut microbiome” inside a narrow transparent tube via the US postal mail service to American Gut. My microbiome made the long trip from Miami to California stuck on a Q-tip. At the time, it seemed that the scientific research into this tiny new world of microbes had come a long way, but to my disappointment, I found out that there is still much to be learned. I found out that my microbiome had not survived the trip intact and had started to bloom, aka “grow new bacteria” during the trip.
In recent years, the microbiome has been featured prominently in the news as one of several paths to personalized medicine. Researchers from diverse fields such as psychology or nutrition or cancer, and many others, are trying to join the race to decipher it. There is great promise in its research, which is why the sector is receiving billions in funding, with major research initiatives in the US and worldwide. But as of today, the microbiome is still a mystery.
A 2014 article titled “20 Things you Didn’t Know About the Human Gut Microbiome” aptly describes the microbiome this way:
The microbiome is defined as all the bacteria, viruses, fungi, archaea, and eukaryotes that inhabit the human body. Collectively referred to as the “second human genome”, the gut microbiome in particular is now being considered a separate “organ” with distinct metabolic and immune activity. The two major areas of microbiota investigation include taxonomic diversity to identify “who” is there and functional metagenomics to figure out what they are doing. There are other human microbiome sites as well, including skin, oral, and vaginal, but the gut is the most popular and diverse neighborhood.
Two years after the article was published it seems that this statement is still true: “Our understanding of the ‘normal’ microbiome patterns, including what constitutes a healthy versus diseased pattern is still in its infancy.” One consensus that emerged back then was that a wide diversity of gut microbiota is a sign of good health, and having only a select few bugs is a sign of bad health. A 2016 article presents some new definitions of the “healthy microbiome”, and they reiterate that a diverse microbiome indicates good health. They point out, however, that it does not hold for all body sites, citing vaginosis as an example. It is interesting to know that people in developed countries have a less diverse gut microbiome which is correlated with chronic diseases and linked to a high-fat, high-refined-sugar, and low-fiber diet. This is useful information for people who care about their health and follow a healthy diet.
As the article points out, one of the biggest outstanding gaps in understanding the basic biology of the “healthy” microbiome is at the level of its molecular function: up to 50% of microbial gene families encountered in the human microbiome remain functionally uncharacterized, even in well-studied environments such as the gut. What that means is that of the microbes we can identify, we don’t know what mischiefs or good deeds they are up to. The article also mentions that most of the attention has been given to bacteria and less to archaea, viruses, fungi, and other microorganisms.
This leads me to my main question: if it is difficult to identify the bugs, and it is difficult to identify what they do, and compounding the problem, the bugs bloom on cross-country trips, then how can a common citizen learn anything useful from his microbiome at this stage of scientific research? The answer to that question requires that we consider a few new developments in microbiome research.
The Blooms: The American Gut Project reported in 2015 that they had received microbiome samples from self-reported healthy people that contained levels of proteobacteria beyond anything previously observed in healthy populations. They determined that these blooms were the result of shipping conditions. They reported that the blooms can be bioinformatically subtracted from the dataset by removing organisms observed to bloom and have promised to publish the method they use to do so. More information has been published in their web site.
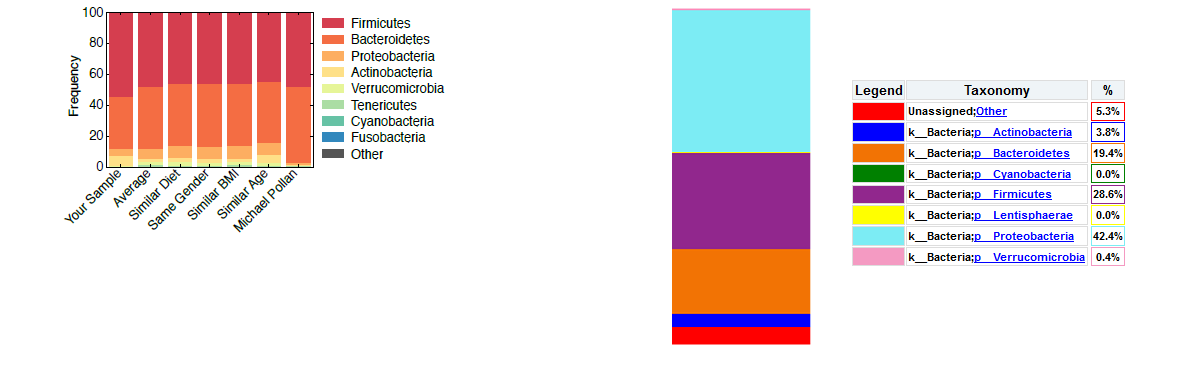
The blooms explain why my results were different to those they reported when I analyzed my own datasets downloaded from the link they sent me. In the image above on the left side, you see the results reported by American Gut. They are not easy to decipher as they are presented in narrow bar charts, with no percentages. The leftmost bar, “Your Sample”, contains my results after they removed the blooming proteobacteria, the other bars are for comparison.
I analyzed my raw dataset, the bacterial 16S rRNA DNA sequences that American Gut makes available, with the NIAID Nephele online tool using a standard bioinformatics pipeline (Qiime). The chart on the right in the top image shows the results. At 42% proteobacteria this dataset contains a very large percentage of blooms compared to the percentage reported by American Gut. The process they use to remove them affects all other percentages, as one can see 28.6% of Firmicutes in the Qiime results which is much less than the approx. 50% shown in the bar chart. Until they publish their method to remove blooming bacteria, it will be hard to reproduce the results.
New handling and storage solutions: The problem with the gut microbiome blooms can be fixed with improved types of specimen collection and storage kits. A 2016 study identified fecal occult blood test (FOBT)) collection kit used for colorectal cancer-screening as a storage method that resulted in the highest 4-days stability without freezing the samples. Freezing the samples also maintains the stability of the microbial populations.
The fascinating handheld or miniature sensors of the future: Recent research by Berkeley Lab used their award-winning PhyloChip, a credit card-sized device, to detect the presence of more than 60,000 species of bacteria and archaea in contaminated water samples. A 2016 study of the mouse gut microbiome published in the Nature journal used a portable nanopore sequencer device, the MinION, to detect bacteria, and found that this miniature sequencer allows for rapid, accurate and efficient determination of the microbial diversity at the species level. Jack Gilbert, a principal investigator at the University of Chicago and the Biosciences Division at the DOE’s Argonne National Laboratory participated in a 2015 roundtable discussion by the Kavli Foundation and came up with a fascinating prediction for the future of nanotechnologies:
We would like to go even smaller, and compress these capabilities into a pill that you could swallow so you could analyze the human microbiome — or metabolome or even the proteinome — at any point in the gut. You could even put an RFID transmitter in there, so the pill could communicate with your phone and you could see what your microbiome was doing in real time.
Self-tracking the microbiome: As reported in 2015, looking at the range of companies currently offering to sequence your genome or microbiome (e.g., American Gut Project, uBiome, my.microbes, DIYgenomics, Genomera), the number of self-tracking projects making use of commercial kits to analyze microbiome is likely to grow in the years to come. As sequencing costs go down, more people are going to perform self-experiments, and new citizen science projects conducted in collaboration with academic labs will gain in popularity.
And so, there is more than one answer to the question about why we should sequence our microbiome now. For now we have to trust that the microbiome/bioinformatics specialist handling your sample will know how to remove the blooming bacteria. Better solutions to improve storing and handling of the microbiome are being implemented, and perhaps we need to research the service we choose before we engage in self-science. As more people get involved in self-tracking it will propel a technological wave of more advanced and smaller portable sensors. Finally, donating our microbiome to research will help advance this scientific field, and if we wait a few years, there will be no need to ship it anywhere. The science and the sensors will have advanced and become miniaturized to such a degree that we can study it right at home, while digesting food with results posted instantaneously to our smartphones.
