Those of us who are curious about the hidden secrets encoded in our genes are excited about the new biomedical technologies that give us access to this genetic information. Our genes can tell a story about who we are. Only a “partial” story because we know that our environment and life style will add to the story. A well-established model of health is represented by this equation: Genome*Exposome=Phenome. The Exposome, is the collection of things we are exposed to, our life style and it has great influence on how we feel overall. Science tells us that we can prevent health problems if we are better informed about our “genetic” health risks, Genome and Genotype information, and adapt our life style to fit our genes. The Phenome and phenotypes are the observed outcomes of the interaction between Genome and Exposome, such as an observed disease, a symptom, a physical change such as weight loss or accelerated heart rate, or a mental state like depression or joy. Doctors, therapists and trainers have been measuring our different phenotypes, and have been monitoring our exposome whenever things go wrong, but they and we have little knowledge about our genetic predisposition for certain phenotypes.
And so, we want to know. At $199, or somewhat less with a discount coupon, the price tag for 23andMe genetic testing is only accessible to few. There are other options that are similarly expensive, among them deCODEme and FamilyTreeDNA offer similar genetic testing services. Those prices are expected to drop as more genetic testing services compete against each other and as sequencing technologies become cheaper. When that happens, more people will benefit from understanding their genes and health risks.
The results from genetic testing are limited by what the company who offers the service is allowed to reveal. In 2013, the FDA intervened, ordering 23andMe to cease and desist from providing analyses of people’s risk factors for disease until the tests’ accuracy could be validated. It took a while, but now 23andme is back in the genetic testing business with FDA approval for certain health related tests, but not all. 23andMe offers 3 Ancestry, 39 Carrier Status, 19 Traits, and 7 Wellness reports. That is not much if you consider that they test 610,500 SNP positions in a person’s genome. A single nucleotide polymorphismus (SNP) is a difference in your genome compared to the majority of people when looking at a single DNA position. SNPs occur normally throughout a person’s DNA. They occur once in every 300 nucleotides on average, which means there are roughly 10 million SNPs in the human genome.
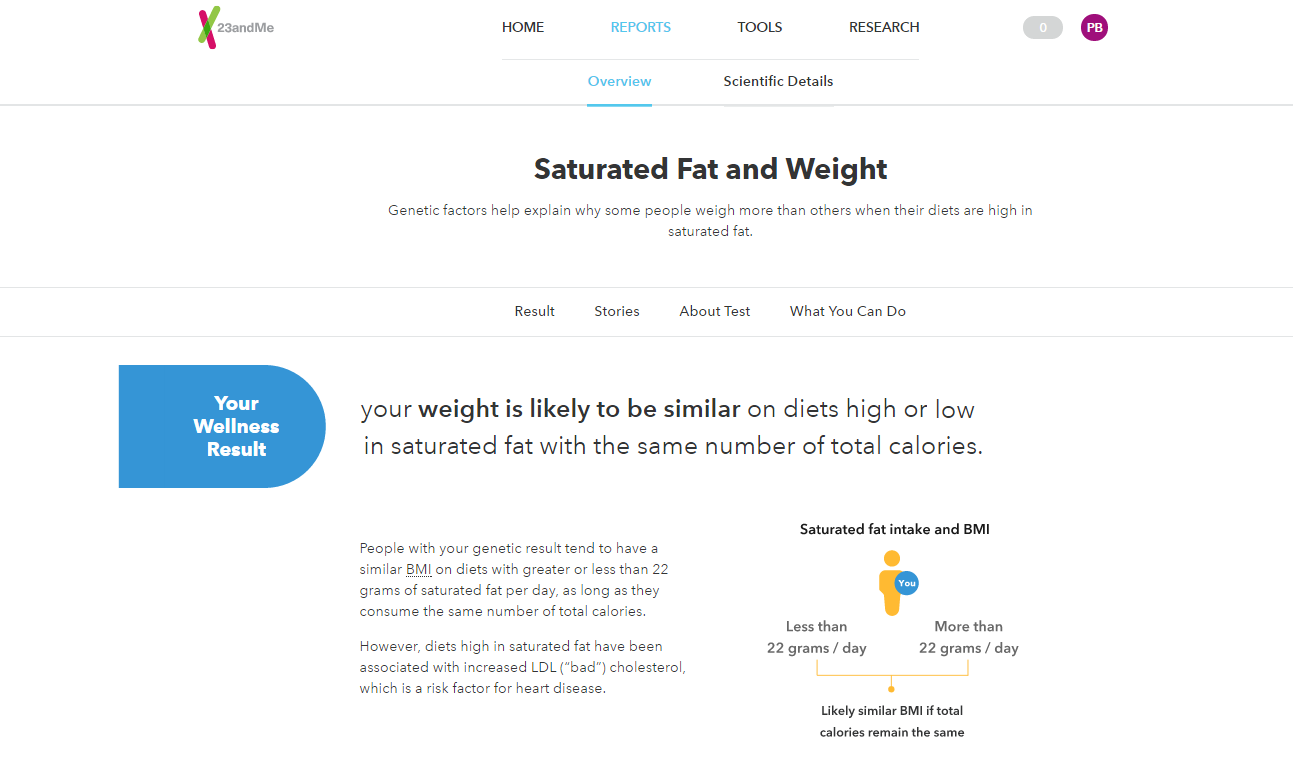
The ancestry reports are cool even when you already know your ancestry. I have always known that I am half Latina, half European, and I am happy to report that my genes confirmed that. The Traits report will tell you what color eyes you have and if you have freckles, among several other traits that you should already know. The Wellness report will tell you if you are lactose intolerant and if you are a restless sleeper among traits you may or may not be aware of. But the interesting aspect of the report is the one that tells you what you don’t know. Yet here, the information is limited. The Carrier Status report is mostly aimed at reporting rare diseases like “Neuronal Ceroid Lipofuscinosis”, and I must admit that I was relieved that my status was “Variant not detected” for all 39 traits. 23andme keeps adding reports, therefore, eventually, there will be more information available and for people who are too busy to do the research themselves, this is their best option.
23andme has a long way to go to get back to reporting the same number of variants they were before the FDA ban. However – both the previous and new 23andMe reports pale in comparison to an analysis of the raw SNP data. 23andMe’s new reports tell you about less than 1% of the health-related variants that are in their raw data, and the raw data is the one that gives you the option to be your own scientist. There are a number of ways to study SNP data and I will list some of them here. I am going to list the study options based on how much you can learn without spending any or very little extra money, but I will not give priority to the easy options because doing your own genetic research should be a fun pastime. With a little guidance, some time to spare, and minimal knowledge of programming or of SNP data, anyone can do it.
SNPedia is my top option because it contains a well curated catalogue of SNP genotype information linked to their phenotypes (i.e. associated disease or trait) and you can access all the data programmatically via Perl or Python. On their web site you can look at information on a SNP by SNP basis. The quickest way to access this information is to add the SNP number at the end of the SNPedia URL, as shown here for rs5082 (a SNP which may influence obesity and heart disease risk): http://www.snpedia.com/index.php/rs5082. With an installation of the free Perl Strawberry and the Perl mediaWiki module, Perl code can be copied from here and here to download about 58,913 SNP genotypes (as of July 2016). After parsing the text it can be saved in a tab delimited file and used in Excel with the Vlookup function to join SNP raw data to the SNPedia results. Once in Excel the data can be sorted, for example by the magnitude, which is a subjective measure of the significance of a genotype but points to the ones who have been studied more. Something that causes confusion is the orientation of a SNP, as the DNA can be read in the forward (+) or reverse(-) direction and this information needs to be taken into account by finding the reverse complement of a reverse/minus SNP.
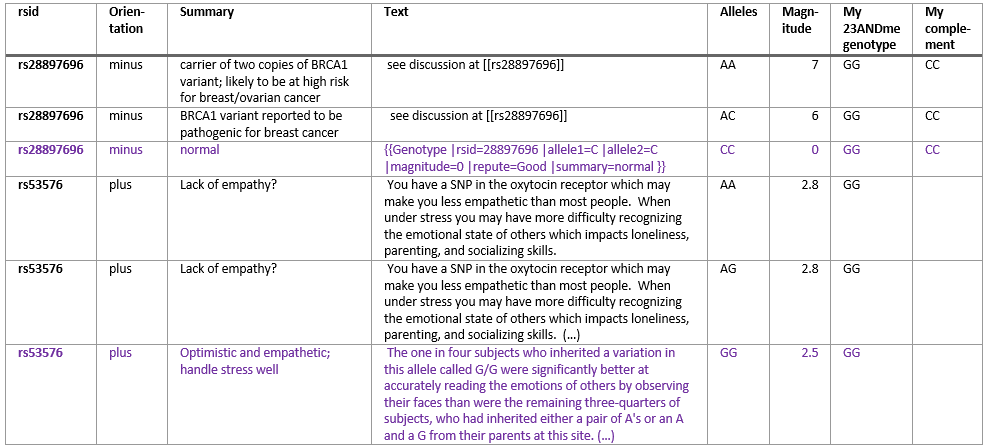
The next option are publicly curated databases that link to dbSNP and GWAS, such as dbSNP itself, ClinVar, and Ensembl, where one can study SNPs, one at a time. In any of these resources, one will find the traits/disease/phenotype association for GWAS SNPs like rs429358 (here in ClinVar, and here in Ensembl), but one will not find them for some SNPedia SNPs, such as rs53576. There are options to connect directly to Ensembl via a MySQL connection and run queries on the variation database. One may also download dbSNP or ClinVar data but finding the appropriate linked data may be more difficult than with the previously discussed methods.The next best option is the GWAS catalogue which contains SNP to trait or disease association based on genome-wide association studies. It is easy to download with no need for programming. The delimited file can be imported into Excel and handled the same way as SNPedia’s data. Like with SNPedia, the orientation problem has to be taken into account to make sense of the data. In this case, an additional complication is that GWAS only reports one “Strongest SNP-Risk Allele” and that this one is often unidentified or “?”. Reading the publication with results of the study sometimes helps but is time-consuming. There is no “Magnitude” score but here one would look at the P-value, and the smaller the number the more significant the association.The next best option which I recommend for people who have little time to do research but would like to read about their SNP information is to pay $5 for a Promethease report, which uses the data from SNPedia. The report shows most of the information that appears in SNPedia but in a nicely formatted, personalized html page and also takes care of the orientation issue. Some information is left out, for example SNP reports on risk alleles of magnitude higher than 4 are not shown. It is also not reported what the other 2 alleles that you don’t have mean which can help keep things in perspective as shown in the table above. Some information could have been left out, for example, it claims that I have an “Increased risk of Male Pattern Baldness”, but their algorithm could have inferred from the absence of Y chromosome SNP variants that I don’t have a Y chromosome.
When looking for other options that allow getting a report for all genotype to phenotype associations, one may consider submitting SNP data to OpenSNP. OpenSNP parses the file with SNPs data and annotates them. However, by signing up for OpenSNP, a person has to declare that he or she has understood the possible risks and side-effects that can occur by making his or her genetical information available because although OpenSNP does not reveal a person’s identity, it makes the genetic data public. If one wishes to share data for health research, OpenSNP should be considered over commercial options, because it uses the data for open science purposes and makes no profit from it. 23andMe will ask you to share your genetic data or survey responses, and 23andMe will sell your data to third parties for their own profit. Before you agree to share your data with companies who make a profit and don’t share the profit with you, you should consider if this process is fair, given that you paid a hefty price to get tested and it is your data.
And then there is Enlis Genomics, who promises that you will “receive an email with a free genome report”, but the report you get is lacking. You get 2 PDF files, one containing a long report for a single phenotype, the other pdf containing 4 other rare “Selected Phenotype Variation Details” (among them “Blood group–lutheran null” which has no directly known disease association). Enlis gives you the option to buy “your annotated genome data” for a price of $39.95.
While there may be other companies or resources available, but these are the most prominent ones. Variations at specific positions in a genome will continue to be studied, and more reports will become available. It is fun to be our own scientist as long as we understand that the science is not cut in stone yet. But our genes, with the exception of cancer, do not change, and we will be able to study our data for a lifetime and learn more as science advances. One can foresee that more genetic testing companies or services may emerge in the near future, but even as they do, new sequencing technologies will allow looking at long stretches of DNA not just a single position. Looking at longer sections of DNA may be a more meaningful analysis of our genetic data, but the price for whole genome sequencing is still too pricey and tools to study our genomes at the personal level are rare. Therefore for now, accessing our raw SNP data from the company that did our genetic testing, purchasing SNP reports or using free SNP resources to study our data are the best options we have to study our personal genetic code.

The degree to which your phenotype is determined by your genotype is referred to as ‘phenotypic plasticity’. If environmental factors have a strong influence, the phenotypic plasticity is high. If genotype can be used to reliably predict phenotype, the phenotypic plasticity is low.